Multiscale Computational Tools for Optogenetics

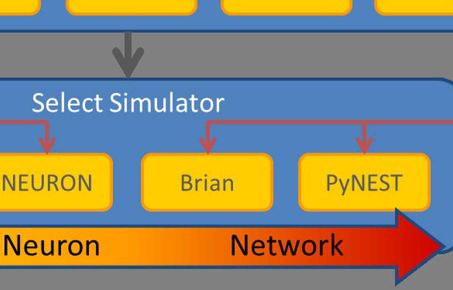
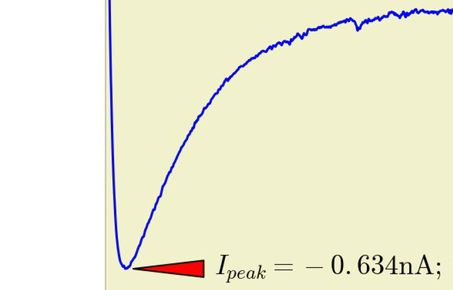
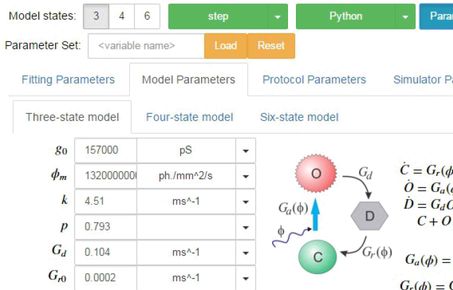
PyRhO is an integrated suite of open-source tools written in Python for parameterising and simulating (rhod)opsins. It provides convenient data structures and routines for fitting kinetic models of opsin dynamics to experimental photocurrents in addition to "ready-made" models of popular opsins. These models may then be simulated in a variety of common stimulation protocols from the individual channel level to whole networks of transfected spiking neurons. PyRhO also comes with a Jupyter notebook based Graphical User Interface (GUI) making it intuitive and easy to use with minimal programming experience.
PyRhO Quickstart
PyRhOmania
How to Cite
If you use PyRhO, please cite our paper:
Evans, B. D., Jarvis, S., Schultz, S. R. & Nikolic K. (2016). "PyRhO: A Multiscale Optogenetics Simulation Platform". Front. in Neuroinform., 10(8). doi:10.3389/fninf.2016.00008
Publications using PyRhO
Please contact us if you cite PyRhO with details of the publication.
How to Contribute
- Fork us on GitHub
- Upload your data
- Suggest improvements
People
People
- Dr Benjamin Evans, the software creator
- Dr Konstantin Nikolic, Principal Investigator
Collaborators
- Dr Simon Schultz, Department of Bioengineering
- Dr Sarah Jarvis, Department of Bioengineering

Funding
UK Biotechnology and Biological Sciences Research Council, grant: BB/L018268/1
UK Engineering and Physical Sciences Research Council, grant: EP/N002474/1
External Collaboration
Please contact us if you would like to collaborate.