Our resources include datasets, protocols, plasmids, and genetic models available for use courtesy of the Beta Cell Genome Laboratory. Expand the relevant sections below for further information.
Accordion widget - Resources
- Islet Regulome Browser
- Dataset downloads
- Plasmids available via Addgene
- Genetic models made by the Beta Cell Genome Laboratory, available via The Jackson Laboratory
- Protocols
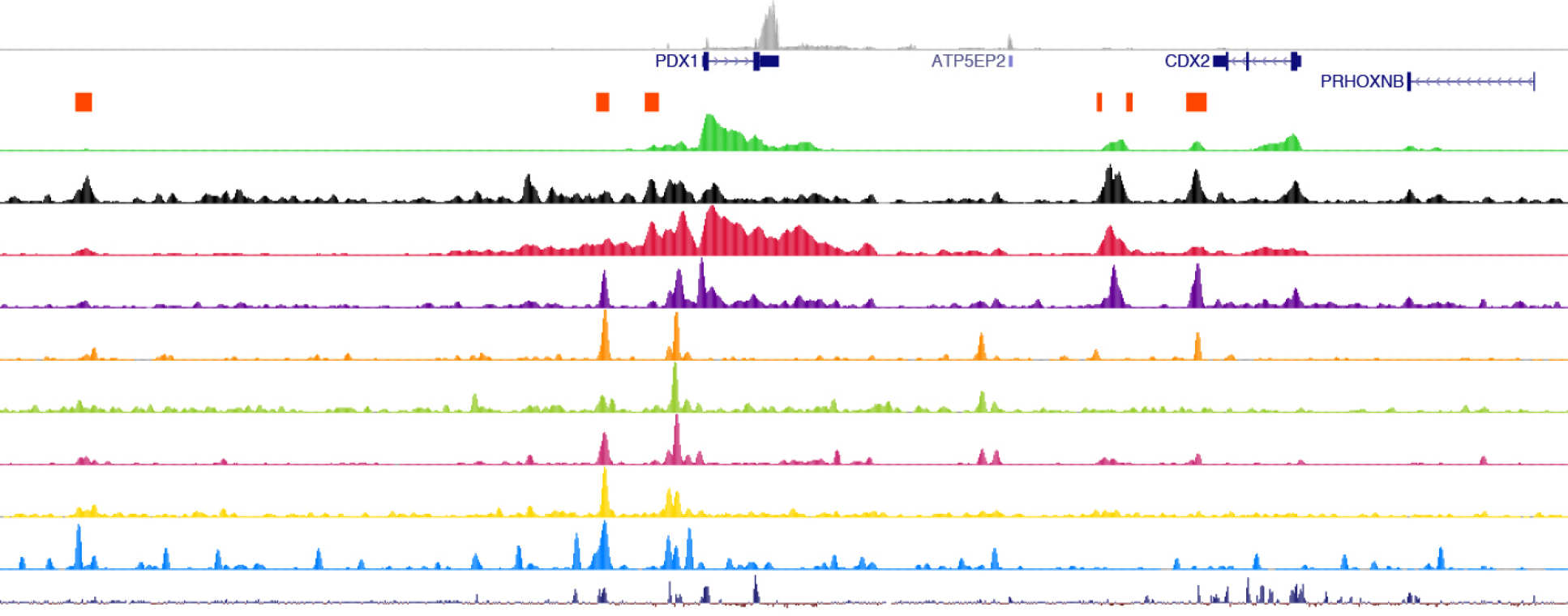
- Genome Browser Tracks
To visualise transcription factor binding sites, DNA-binding motifs, active enhancers and promoters in human pancreatic islets and embryonic pancreas, please visit the Islet Regulome Browser generated by Loris Mularoni, Lorenzo Pasquali. This Browser is developed and maintained by the Pasquali lab at the Institute Germans Trias i Pujol, Badalona. We recommend using Chrome, Safari or Firefox for optimal functionality.
1. Morán, Akerman et al., Cell Metabolism 2017: Human β Cell Transcriptome Analysis Uncovers lncRNAs That Are Tissue-Specific, Dynamically Regulated, and Abnormally Expressed in Type 2 Diabetes
BED12 file of the location and exon annotations of the human pancreatic islet long non-coding RNAs, in hg18 coordinates. Note that the "overlapping antisense" lncRNAs are not included in this list, as they are already part of other gene annotations: Islet LncRNA coordinates.bed12 [Excel]
All RNA-seq and ChIP-seq datasets used in this study can be found at ArrayExpress, accession number E-MTAB-1294.
2. van Arensbergen et al., Genome Research 2010: Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta cells to adopt a neural gene activity program
COD files as output by Cisgenome HMM analysis on Affymetrix mouse promoter arrays. These files contain the enriched intervals (mm8/NCBI36) for H3K4me3 and H3K27me3 for all analysed cell types: part1 [ZIP], part2[ZIP].
3. Gaulton, Nammo, Pasquali et al., Nature Genetics 2010: A map of open chromatin in human pancreatic islets
FAIRE open chromatin sites identified in human islet: FAIRE open chromatin sites [Excel]. This file provides genomic coordinates for all consistent sites across the reference sample used to chart our first draft of human islet open chromatin map and an additional biological replicate of similar purity.
Raw and processed ChIP-seq data used in this study can be found in E-MTAB-1919.
Human islet FAIRE-seq raw data, alignments in the form of mean-centered base counts, and enriched sites at three thresholds for all samples are found in GEO GSE17616, E-GEOD-17616 (where samples 1 and 2 are replicates of HI32 and sample 3 refers to HI10) and E-MTAB-3199 (where all data refers to HI32). HI10 data and replicate 1 of HI32 are of better quality and are recommended for analysis of human islet open chromatin landscape.
Microarray data generated by hybridization of islet FAIRE preparations is also available in GEO GSE17616.
4. Pasquali, Gaulton, Rodríguez-Seguí et al., Nature Genetics 2014: Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants
In this study, we combined ChIP-seq of chromatin marks and key islet transcription factor with RNA-seq in human islets to map cis-regulatory networks in this primary tissue. The output of this paper provides a reference map to dissect genetic variants that alter the susceptibility for Type 2 diabetes and assist efforts to generate new beta-cells by transcriptional programming strategies.
Raw and processed files of the ChIP-seq data are available at ArrayExpress under accession number E-MTAB-1919 (Pancreatic islet epigenomics reveals enhancer clusters that are enriched in Type 2 diabetes risk variants).
RNA-seq data used in this study under E-MTAB-1249 (RNA-seq of several species pairs of domesticated and wild animals to investigate brain gene expression patterns).
Excel files with genomic coordinates of C1-5 sites related to Fig2a: Genomic coordinates of C1-5 sites [Excel]
Clustered C3 sites related to supplementary Fig6 (hg18 build): Genomic coordinates of clustered C3 sites [Excel]
5. Cebola, Rodríguez-Seguí, Cho, Bessa, Rovira et al., Nature Cell Biology 2015: TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors
In this study, we mapped the regulatory landscape of embryonic multipotent progenitor cells (MPCs) that give rise to all pancreatic epithelial lineages. Using human embryonic pancreas and hESC-derived progenitors, we performed RNA-seq, as well as ChIP-seq for histone modifications and transcription factors that are key regulators of pancreas development. This paper provides a resource for the study of embryonic development of the human pancreas, enabling the mapping of genetic variants associated with developmental defects of the pancreas, and supporting efforts to generate new pancreatic progenitors and beta-cells by in vitro transcriptional programming strategies.
Summary of the study at Imperial College website: "Pancreas cells' genetic instructions open doors to diabetes treatment".
RNA-seq data from this study are available at ArrayExpress Archive under E-MTAB-3061 (RNA-seq of in vivo and in vitro MPCs).
Raw files of the ChIP-seq data are available at ArrayExpress Archive under accession number E-MTAB-3061 and E-MTAB-1990.
Excel file with core MPC genes (expression and Z scores provided for in vivo and in vitro MPCs): Core MPC genes [Excel].
Excel file with genomic coordinates of MPC enhancers (Figure 2): MPC enhancers (hg18 build) [Excel].
Excel file with genomic coordinates of MPC cis-regulatory modules (Figure 4): MPC cis-regulatory modules (hg18 build) [Excel].
Excel file with genomic coordinates of clusters of MPC cis-regulatory modules: Clusters of MPC cis-regulatory modules (hg18 build) [Excel].
6. Miguel-Escalada, Bonàs-Guarch, Cebola et al., Nature Genetics 2019: Human pancreatic islet 3D chromatin architecture provides insights into the genetics of type 2 diabetes
Datasets from this paper have been uploaded on the Ferrer Lab CRG website, DATASETS.
Hnf1bCreER - This model may be useful for generating specific targeted mutations or fate-mapping studies of Hnf1b expressing cells.
Ins1Cre - Cre recombinase is expressed in insulin-producing beta cells of the pancreas.
Ins1CreER - Cre-ERT2 is expressed in insulin-producing beta cells of the pancreas. This model is useful for applications requiring tamoxifen-induced deletion of floxed sequences in beta cells.
The Beta Cell Genome Regulation Laboratory is currently using a range of protocols, available for download via the following links:
Our Human Islet LncRNA annotations are available as a public ‘trackhub’ called "Human Islet lncRNAs" for visualization in the UCSC genome browser.
- The trackhub can be directly visualized with this link.
- Alternatively: you can load the trackhub directly from UCSC using the name Human Islet lncRNAs
- Description of the hub

Links
General enquiries
Beta Cell Genome Regulation Lab
ICTEM Building, 5th floor
Hammersmith Campus
Du Cane Road
London
W12 0NN
Enquiries
d.cieslak-jones@imperial.ac.uk
+44 (0)20 7594 2739


