Probing cells' recycling centres reveals new drug targets
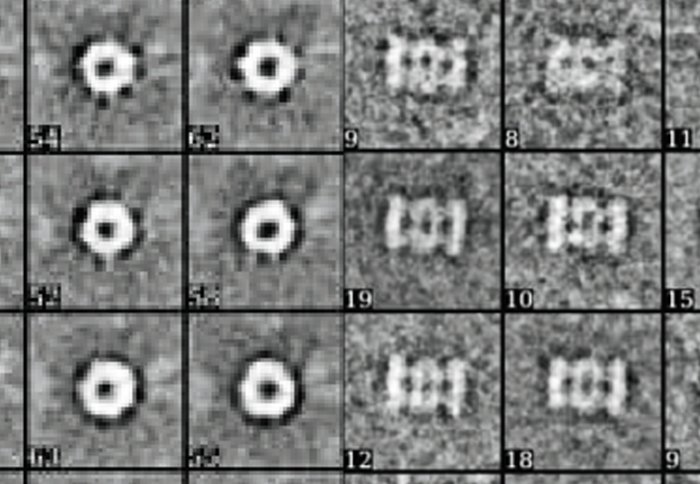
Top and side views of proteasome chambers. Image courtesy of Dr Elmar Behrmann.
Scientists have found differences in the ways cells break down proteins, revealing mechanisms that could be targeted in diseases such as cancer.
Proteins come in many shapes and sizes and perform a staggering array of roles within each individual cell in our bodies. Cells routinely break down unwanted proteins and recycle them into new proteins according to the cells’ changing needs.
Proteasomes are the cells’ recycling centres, breaking down unwanted proteins. They will also break down proteins from foreign bodies in the cell, such as viruses and bacteria.
Protein recycling is one of the most promising routes in looking for new drug therapies.
– Professor Michael Stumpf
This process plays a role in the body’s immune response to infection. As proteasomes chop up proteins, they send out small fragments that can be interpreted by immune cells to determine whether an invasion is in progress.
However, these fragments must be at a certain length with certain characteristics in order to be identified, which is especially well done by specialised immunoproteasomes.
Dying in their own garbage
Now, a group of researchers led by Imperial College London have discovered differences between the ways standard proteasomes and immunoproteasomes handle proteins. This could help scientists design drugs that inhibit the function of immunoproteasomes but not standard proteasomes.
Some types of cancer have more immunoproteasome than standard proteasome compared to healthy cells. Researchers have speculated that if they could target the immunoproteasomes and slow the cells’ recycling function, then this could help to tackle these certain types of cancer.
Tumour cells grow and divide much faster than healthy cells, meaning their proteasomes also need to work fast to ensure all the extra proteins are recycled.
Inhibiting this recycling mechanism would mean cancer cells cannot get rid of old proteins fast enough and ’die in their own garbage’, according to Professor Michael Stumpf from the Department of Life Sciences at Imperial.
Drugs that inhibit specific functions of immunoproteasomes, essential for tumour cells, could target some types of cancer while having minimal impact on healthy cells.
Probing proteasomes
Proteasomes have central chambers where the cutting up of proteins actually takes place, but also doors to the chamber that control when and how proteins can enter and exit.
Dr Juliane Liepe and Professor Michael Stumpf from the Department of Life Sciences at Imperial partnered with colleagues at the Universitätsmedizin Berlin in Germany to understand more about what opens these doors and how fast proteins pass through them. Their work was published last month in the journal eLife.
The team modelled the possible mechanics of protein transport into the proteasomes on a computer, and then used experiments with human proteasomes in the lab to validate the results. They found that standard and immunoproteasomes had different ‘buttons’ that opened the chamber, and that proteins entered and exited at different speeds.
Full of promise
Professor Stumpf explained the importance of their approach in creating drugs for cancer: “To design inhibiting drugs for these proteasomes, we have to account for the buttons and not just the box – the mechanisms that allow proteins into the chamber and not just the structure of the chamber itself.”
“It is important to get the transport step correct when understanding the process – but this has largely been ignored in favour of looking at the cutting part,” added Dr Liepe, the lead author of this study.
Professor Stumpf adds: “Protein recycling is one of the most promising routes in looking for new drug therapies that could increase or decrease rates of recycling to control protein levels in the cell.”
-
'Quantitative time-resolved analysis reveals intricate, differential regulation of standard- and immuno-proteasomes' by Juliane Liepe, Hermann-Georg Holzhütter, Elena Bellavista, Peter M Kloetzel, Michael PH Stumpf and Michele Mishto is published in eLife.
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Hayley Dunning
Communications Division