Nanoscale images reveal bacteria motor parts in unprecedented detail

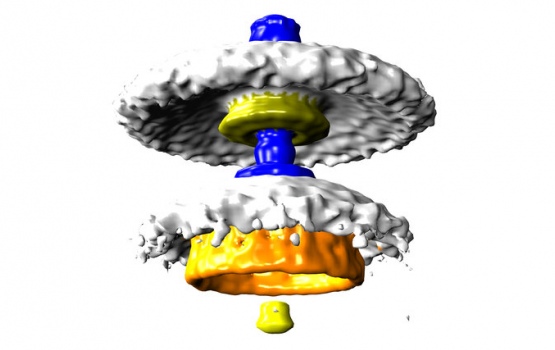
Three bacterial motors. Units of torque are piconewton nanometres (pN nm).
Nanoscopic 3D imaging has revealed how different bacteria have geared their tiny propeller motors for a wide range of swimming abilities.
Many bacteria swim using flagella – long tails that are attached to tiny motors made of proteins, just tens of nanometres wide. These motors spin the flagella, which work as nanoscale propellers to drive the bacterium forward.
For the first time, we have been able to see and explain how these nanoscale molecular machines have evolved in bacteria to colonize new environments.
– Dr Morgan Beeby
Despite motors in diverse bacteria having the same core structure, different bacteria vary widely in their swimming power. For example, Campylobacter jejuni, which causes food poisoning, can swim powerfully enough to bore through the mucus that lines the gut, an environment too thick and sticky for other bacteria to push through.
The reasons for these differences in swimming ability have remained obscure – until now. Using a recently installed high-powered electron microscope at Imperial, a team of researchers lead by Dr Morgan Beeby from the Department of Life Sciences has been able visualize these motors in unprecedented detail.
Their visualizations of these motors explains the differences in swimming ability, mathematically accounting for differences in motor power. The work is published in Proceedings of the National Academy of Sciences.
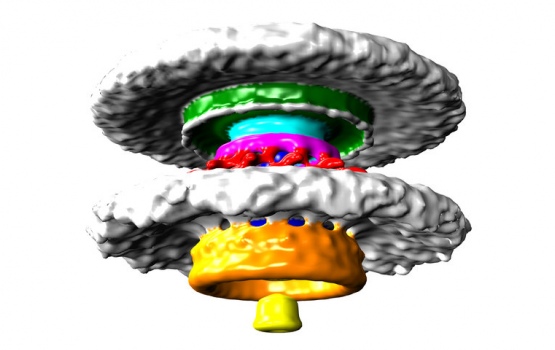
Bacterial flagellar motors work on a rotational mechanism, spinning their long flagellar tail to produce a helical propeller-like motion. The team found that stronger swimmers have evolved by adding extra parts to their motors, making more powerful motors that have increased turning force, or torque.

Lego model of bacterial flagellum
In flagellar motors, the turning force is produced by a ring of structures called stators around the motor. The team found that Campylobacter jejuni had almost twice as many stators positioned around the motor than in Salmonella, and that these structures sat in a wider ring. More stators provide increased torque, and the increased width of the ring means individual stators exert more leverage when rotating the helical propeller.
However, not all bacteria need to be so powerful and swim through such viscous environments as stomach mucus. Instead, another bacterium the team looked at, a close relative of Vibrio cholerae, the bacterium that causes cholera, has evolved a motor with only intermediate power.
“For the first time, we have been able to see and explain how these nanoscale molecular machines have evolved in bacteria to colonize new environments,” said Dr Beeby. “It’s a fascinating insight into the awe-inspiring diversity of life that has evolved on Earth, and also presents possible drug targets. We may be able to design drugs that specifically sabotage the flagella only in targeted bacterial species.”
The team used a method called electron cryo-tomography to rapidly freeze the bacteria to -180?C. This prevents ice crystals from forming that would break the structure apart, allowing the researchers to image the flash-frozen motor from all angles and build up a 3D model.
By looking at distantly related bacteria from different branches of the evolutionary tree, the team speculate that the ability to alter torque in this way may have evolved up to two billion years ago.
“Entire branches of the bacterial family tree have evolved motors with different torques, leading to a diversity of species each geared to their own environment,” said Dr Beeby. The team is now investigating how and when the evolutionary steps that altered motor torque happened.
-
'Diverse high-torque bacterial flagellar motorsassemble wider stator rings using a conservedprotein scaffold' by Morgan Beeby, Deborah A. Ribardo, Caitlin A. Brennan, Edward G. Ruby, Grant J. Jensen,and David R. Hendrixson is published in Proceedings of the National Academy of Sciences.
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Hayley Dunning
Communications Division