A tale of two specialities: how two researchers combined to solve a problem

A clinician and non-clinical researcher have combined forces to investigate the cause of several patients presenting with the same condition.
Dr Phillip James and Professor Paul Cullinan of the National Heart and Lung Institute recently presented a case study of how their two fields of expertise have combined to help patients. The patients worked in an aircraft’s part factory as metal turners, making metal components, and presented with hypersensitivity pneumonitis (HP) which is an inflammation of the walls of the air sacs in their lungs.
Dr James explains how this collaboration came about and what they discovered.
Identifying the outbreak
The first patient presenting with the condition (known as the ‘index’ case, that is the first recognized case in a group of related cases) was identified when a patient was referred to Professor Paul Cullinan. The patient had abnormal lung function scores and anomalies in a chest radiograph. Paul noted he was working as a metal turner and was aware of HP in metal turners. The condition generally occurs in spatio-temporal clusters as an “outbreak”. It is thought that something living contaminates the metal working fluid, and that inhalation results in an allergic reaction in people exposed to high levels of the fluid. The fluid itself is aerosolized (converted into a fine spray) through the cutting process, and without adequate protection can be inhaled by nearby workers. By chance at a teaching session Paul was giving to trainees, another respiratory consultant realized that one of her patients had very similar symptoms and asked Paul for some ideas. It turned out that this patient was the index case patient’s supervisor at the same factory and worked next to him on the machine line. Paul and his team organised a detailed survey of the several hundred workers at the factory which resulted in the identification of three further patients, all of whom worked in the same corner of the factory. They each had work-induced HP (also known as extrinsic allergic alveolo-bronchiolitis).
A meeting of minds (and research)
Previous evidence suggests that microorganisms do live in metal working fluid and attempts to confirm this have included trying crude culture methods to detect living organisms. There is some previous evidence to suggest that a particular group of microorganisms may be causing this disease. They are known as the mycobacteria and are of the same family as that which causes tuberculosis (TB). I had done some previous work on NTMs (non-tuberculous mycobacteria) meaning the ones that don’t cause TB, and their distribution in the environment, as well as molecular based diagnostics of TB in badgers. Paul heard of this during a talk I was giving and wondered if we could use modified versions of the molecular methods I had developed to try to find out if there were any NTM in the metal working fluid samples he had taken from the factory machines. Paul gave me 33 numbered vials of metal working fluid which meant I didn’t know anything about the factory layout (i.e. where the samples came from) or where the outbreak was; everything was carried out in a ‘blinded’ fashion. The challenge he set me was to try to work out where the outbreak occurred.
Investigating what lies beneath
Mycobacterium
First off I just wanted to discover if I could actually see anything which looked like mycobacteria in the samples. I used series of dyes which stain a particular component of the mycobacterial cell wall (the mycolic acids) and wouldn’t be taken up by other organisms. Mycobacteria are rod shaped and approximately 2μm in length. After the staining I saw twoμm rod-shaped organisms which had taken up the particular dye for mycobacteria. The next job was to try to find out what this organism was.
To achieve a taxonomic identification of the organism I amplified and sequenced a region of the mycobacterial genome. This part of the genome is unique to all mycobacteria but its sequence is different for each species of mycobacteria i.e. Mycobacteria tuberculosis and Mycobacteria abcessus will both have the gene but the sequence of bases will be different. I put the sequences from the metal working fluid into a phylogenetic tree with sequences from known mycobacteria. All the sequences from the factory were identical to Mycobacteria avium giving us a robust identification of the mycobacteria in the samples. The next job was to see if there were different amounts of the bug in different areas and whether the actual amount of the organism related to the outbreak area.
To detect the DNA sequence of the bug in question in the samples I used a quantitative polymerase chain reaction (PCR) procedure. This allowed us to work out how many copies of the gene were in each sample effectively telling us how many mycobacteria were in a millilitre of metal working fluid per machine.
The results are in
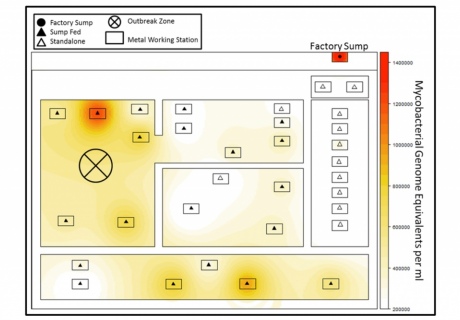
Image showing map of the factory floor showing that particular machines in the outbreak zone had high levels of the mycobacteria in question.
It turned out there was in excess of 1.4 million Mycobacteria avium organisms in samples from the factory sump and over 1.2 million in a single machine right at the heart of the outbreak zone. Although it looks like the sump is a focal point for contamination spreading these organisms, the total bacterial burden does not associate with the outbreak zone.
We are now in the process of publishing a report of this study and working on analysis of UK samples with the Health & Safety Executive (HSE). In the future we would like to consider the point of use immunoassay perhaps with clinical applications and consideration of metal working fluid management systems.
The Grand Rounds are run by the Royal Brompton and Harefield NHS Foundation Trust (Brompton campus) on Thursdays at 1pm; see their webpage for more details.
Image credit: Mycobacterium tuberculosis Bacteria by NIH Image Gallery used under Creative Commons license.
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Phillip James
National Heart & Lung Institute
Ms Helen Johnson
Communications Division