Bacterial machines that help create defensive 'mats' mapped by researchers
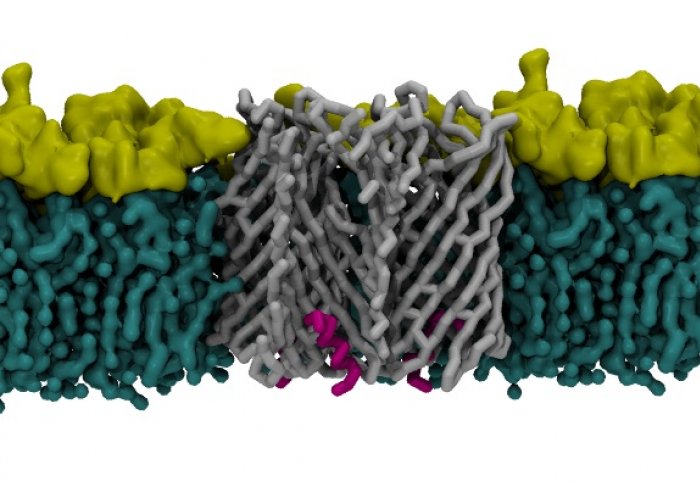
A nanopore through which amyloid proteins are secreted
The way that some bacteria produce the materials that form 'biofilms', which help them evade antibiotic attack, has been uncovered by scientists.
The latest finding could eventually lead to new ways of getting around antibiotic resistance and help in making new, biologically inspired nanomaterials.
When bacteria are stressed, for example when attacked by the immune system or antibiotics, they can club together and form a biofilm. Biofilms can also affect water quality and ‘foul’ surfaces in industrial settings, damaging their performance.
Biofilms make bacteria more resistant to antibiotic attack and discovering more about their assembly will open new avenues for tackling antibiotic resistance.
– Professor Steve Matthews
Bacterial biofilms are reinforced with tough protein fibres called amyloids. Uncontrolled amyloid growth in humans can affect health – for example, amyloid plaques associated with Alzheimer’s disease.
However, bacteria can control amyloid protein and fibre growth. Understanding these processes is therefore an interesting avenue of research to aid the search for new ways of preventing amyloid disease in humans and fouling by biofilms.
Now, researchers led by Imperial College London have mapped the machinery that one group of bacteria use to secrete amyloids. The study, on Pseudomonas bacteria, is published today in Nature Communications.
Lead researcher Professor Steve Matthews, from the Department of Life Sciences at Imperial, said: “Biofilms make bacteria more resistant to antibiotic attack and discovering more about their assembly will open new avenues for tackling antibiotic resistance.
“For example, drugs aimed at interrupting the bacteria’s ability to secrete amyloids would render bacteria more susceptible to antibiotic treatment.”
After the amyloids are manufactured inside the cell, bacteria then shuttle them through pores in their cell membranes. The team determined the structure of the Pseudomonas amyloid pore and revealed a ‘gate’ that must be opened and closed to allow the amyloids out. It is this mechanism that the team plan to study in more detail.

Multitude of applications
Besides applications in medicine and manufacturing, research on amyloid formation and membrane pores has applications in nanomaterial fabrication and biosensing.
Co-author of the study Dr William Hawthorne, from the Department of Life Sciences at Imperial, said: “Amyloids are some of the strongest fibres in nature, for example spider silk contains amyloid-like structures. We can use synthetic biology techniques to produce fibres decorated with new features.
“Understanding more about how bacteria make the fibres means we can get closer to creating new, strong nanomaterials with tailored properties.”
Bacterial cell membrane pores are also interesting to researchers trying to analyse biological molecules. As molecules passes through the pores a unique signature is generated that can be used to identify the molecule in question, such as in DNA. Researchers could use their knowledge of how these pores work in nature to improve their performance as a biosensors.
The study, which was funded by the Wellcome Trust, involved researchers from the University of Oxford, Duke-NUS Medical School in Singapore, Aarhus University in Denmark, Aalborg University in Denmark and a pupil from the Walthamstow School for Girls in London, who was doing a placement in the Imperial lab.
-
‘A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis’ by Sarah L. Rouse et al. is published in Nature Communications.
Article supporters
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Hayley Dunning
Communications Division