Nobel Prize-winning technique sheds new light on DNA replication
by Ryan O'Hare
DNA double helix
An imaging technique used to reveal the molecular detail of structures has helped scientists to shed new light on how organisms copy their DNA.
The international team, which included Imperial scientists, used cryo-electron microscopy – the technique behind this year’s Nobel Prize in Chemistry – to reveal the structure of a key enzyme involved in the earliest steps of the DNA replication process.
DNA replication is fundamental to all living cells, enabling them to divide for growth, but mistakes in this carefully orchestrated process can be catastrophic for cells and lead to devastating disease.
We have managed to reveal another very important piece of the puzzle of how cells make new copies of their DNA
– Dr Christian Speck
Institute of Clinical Sciences
By studying the structure of the enzymes behind the process and which carefully unravel and copy DNA, scientists hope to shed new light on the process.
The researchers say the findings, published in the journal Proceedings of the National Academy of Sciences, could lead to new insights into the basis of cancer and genetic disease.
The international team, led by Imperial College London, the Cold Spring Harbour Laboratory and the Van Andel Research Institute, studied one of these enzymes, called Mcm2-7
Professor Christian Speck, from the Institute for Clinical Sciences at Imperial, said: “We have managed to reveal another very important piece of the puzzle of how cells make new copies of their DNA. Helicases are crucial to this fundamental process, and techniques like cryo-EM are enabling us to capture a much clearer image of what they look like and how they work.”
Duplicating DNA
Before replication can begin, two enzymes called helicases form a single unit on the DNA double helix. When they split to form two functional enzymes, they pull the two strands of the double helix apart, like a gap appearing between the threads in the middle of a piece of rope.
As the DNA replication process begins the enzymes then move along the DNA in opposite directions, ‘unzipping’ the double helix as they go, with the new strands filling in behind them, a mirror image of the exposed strands. The result is two new DNA helixes, comprising of one half of the original strands and a newly made strand.
Using cryo-EM, the team was able to image these helicase enzymes, called Mcm2-7, revealing its 3D structure, illuminating how the enzymes latch on to 60 base pairs or letters of the DNA.
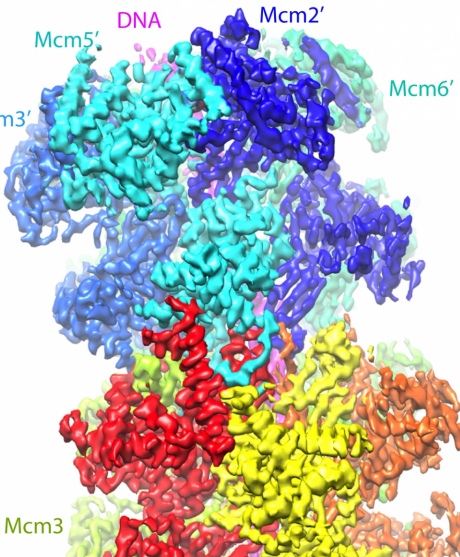
A computer-generated model of the McM2-7 enzymes binding to the DNA double helix
From computer rendering of the structures, the team think that they now understand how these enzymes kick off the whole process. As the helicases form around the DNA double helix they cause it to kink into a zigzag shape which puts pressure on it.
According to the researchers, the effect is like a spring-loaded mechanism when the enzymes separate, pulling the DNA apart.
Huilin Li, a professor at the Van Andel Research Institute and senior author of the paper, said: “These are processes at the very foundation of life that have largely remained a mystery to biologists since the discovery of DNA double helix more than 60 years ago.”
Professor Speck added: “Thanks to this Nobel prize winning cryo-EM technology, we can now directly visualize the operational mechanism in action, for the benefit of research and to improve health for people around the world.”
-
‘Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model’ by Noguchi, Y et al. is published in the journal Proceedings of the National Academy of Sciences.
This article is based on materials provided by the Van Andel Research Institute.
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Ryan O'Hare
Communications Division