How do cells connect with their neighbours?
An Imperial College London team, led by Dr Vania Braga of NHLI, have developed a way to take a closer look at proteins involved in cell-cell contact.
As humans we are made up of lots of different cells that carry out a multitude of different tasks. Our evolution from simple uni-cellular organisms to our complex form today has allowed us to adapt to different environments and evolve. A key aspect of multi-cellularity is cell-cell adhesion, the process by which cells interact and attach to their neighbouring cells, which is carried out by the molecules which make up the cell surface. This cell-cell adhesion allows cell specialization, to carry out different tasks in the body, because it is needed for tissue integrity and processes such as cell division. However to date there is little understanding of how the molecules in the cell walls work together to keep the cell adhesion.
Skin cells (epithelial cells) need to be extra tight with each other as they have to sustain mechanical stress and create a barrier between us and our environment. If you stop this property of tight cell cohesion, the cells can take on another identity, potentially leading to cancer progression. How do different diseases target cell-cell contacts and how do they disrupt them?
"What excites me about it is there is an integrated approach, things are all inter-related"
– Dr Vania Braga
Study lead
A team from Imperial College London, led by Dr Vania Braga of NHLI, have developed a new technique to allow us to visualise and then group the many proteins that regulate the cell-cell junctions. This collaboration between biologists and bioinformatics led to the creation of a quantitative measurement tool to look at these proteins in a new light. This resulted in the identification of 156 proteins that form the cytoskeleton and regulate cell-cell adhesion.
E-cadherin is known to play an important role in cell adhesion, when it becomes dysfunctional the cells are unable to attach strongly to each other or sustain stress. E-cadherin adhesion between neighbouring cells is a complex event involving a variety of cellular processes. However we don’t know how these processes are coordinated. E-cadherin works with the cytoskeleton made of actin filaments and a number of other proteins that cling onto the actin structures and modulate their organization. Yet, how they interact with other cytoskeletal proteins to form cell-cell junctions is again unclear. The investigation of these processes had been hindered by a lack of a quantitative method for analysis.
In their work Vania Braga’s team collaborated with mathematicians to develop computer-vision software to read images and categorise cell junctions in various states of cohesion. They made stains and taught the computer to recognise the border of cells, which it can then measure, and used an algorithm to find mathematical patterns. This provided a way of quantifying the state of the cell contacts. By removing each protein one by one, the researchers could see the effect on the cell junctions and find out which proteins are necessary to maintain contact. The proteins were then grouped by the way they disrupted the junctions – called ‘phenotypic clustering’. This approach allowed the scientists to identify new cytoskeletal proteins (such as crosslinkers and signalling molecules) working at cell-cell contacts, their functional hierarchy and how their pathways modulate E-cadherin adhesion. The prediction of which proteins may work together can be useful for designing therapies, e.g. for a patient requiring treatment, drugs can be used to inhibit another protein in a process rather than a core, essential protein. This may reduce side effects that occur when the core protein is treated, but still allow the required process to be affected, resulting in a better outcome for the patient.
The novelty of this work has been to bring mechanistic, functional and hierarchical insights to the way cells adhere to each other. Recent work from the lab shows precisely the interlinked nature among proteins. The signalling molecule CdGAP is up-regulated (increasing its response) in different cancers and disrupts cell contacts. However, when the cytoskeletal protein Ajuba interacts with CdGAP, it prevents the destruction of cell-cell adhesion by keeping CdGAP dormant at cell to cell adhesion. The researchers are now looking for suggested interactions between proteins from the groupings they have found and then testing if these proteins do interact in vitro and in cells . There is also a potential impact in other tissues in which cell-cell adhesion plays a prominent role such as cardiomyocytes adhesion in the heart or endothelial cells in veins and arteries.
Finally, the new software developed allows the researchers to cope better with the large amount of data processing now required, through the analytical power of maths. Such tools allow labs to move much further with their biological research as analysis is now much faster. This ties in with the general leap forward we have seen in laboratory machinery that has progressed us exponentially to obtaining numerous measurements at a time. With more results to process, we require new techniques, but these large datasets we are creating can be explored further in the future, from other angles.
--------------------------------------------------------------------------------
With thanks to Chris Tomlinso (Bioinformatic Support, Faculty of Medicine), John Lees (Bioinformatics, UCL), and our Masters and PhD students. And our funders BHF Centre of Research Excellence, Cancer Research UK, Medical Research Council, BBRSC and Wellcome Trust.
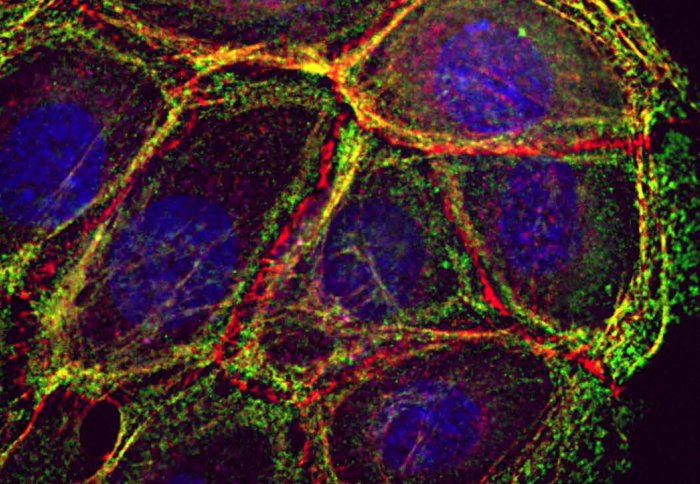
Photo caption: Figure shows staining of epithelial cells stained with phalloidin to reveal F-actin (red) and phosphorylated regulatory myosin light chain (green) to show contractile bundles of actin. Nuclei appear in blue. F-actin accumulates at cell-cell contacts where it co-localise with E-cadherin (not shown), in thin bundles parallel to junctions and lamellae protrusions.
Article supporters
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Ms Helen Johnson
Communications Division