New tissue technique gives stunning 3D insights into the human brain
by Ryan O'Hare
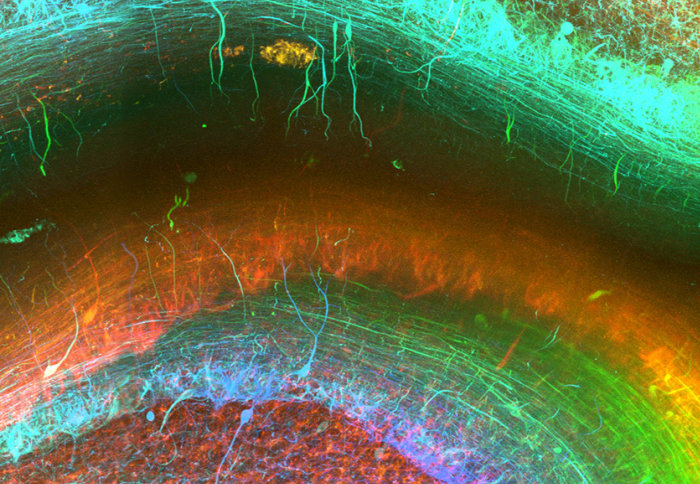
Imaging method enables researchers to generate stunning 3D images of human brain tissue
Imperial researchers have helped develop a breakthrough imaging technique which reveals the ultra-fine structure of the brain in unprecedented detail.
The next generation method enables researchers to generate 3D images of fresh and archived brain tissue samples, resulting in stunning images of the human brain at the microscopic level.
Developed through an international collaboration between scientists at Imperial College London and The University of Hong Kong, the researchers believe the technique could help to shed new light on the basis of neurological diseases which affect millions around the world.
Traditionally, imaging brain samples has involved taking small tissue samples cut into ultrathin slices which can be stained to reveal characteristics of interest, such as proteins or other markers associated with disease.

In recent years, however, advances in molecular tagging techniques and the availability of laser microscopes have led to the development of modern tissue clearing techniques.
These techniques make brain tissue transparent and enable researchers to see the anatomical structure in 3D. However, these techniques were originally developed for rodent brain tissues and there have been very few studies on human tissue.
By using tools such as these in the lab we will be able to visualise how cells interact with each other in 3D Professor Steve Gentleman Scientific Director of the Parkinson's UK Brain Bank at Imperial
Many of the problems centred around the unique properties of the human brain, as well as preservation and processing of human tissue at post mortem.
To overcome these problems the team, led by 3 medical students, have developed a new tissue clearing solution, OPTIClear, enabling a wide range of molecular labelling methods for 3D visualisation of fresh and archival human brain tissue.
Using this new approach, they have been able to stain nerve cells, glial cells and blood vessels, as well as pathological markers such as tangles of tau protein found in the brains of Alzheimer’s patients, in exquisite detail and determine how they relate to each other in 3D space.

“These techniques enable us to reveal the microscopic structure of the human brain in spectacular detail,” said Professor Steve Gentleman, Scientific Director of the Parkinson's UK Brain Bank at Imperial College London.
He added: “By using tools such as these in the lab we will be able to visualise how cells interact with each other in 3D and learn more about the pathways and connections that are damaged in the common neurodegenerative brain conditions which have such an enormous impact on people’s lives.
“This work is only made possible by the wonderful altruism of our brain donors and their families”


According to the researchers, the method is relatively inexpensive, is time efficient and easy to carry out, and is likely to form the basis for further technique development.
It is hoped that a better understanding of the connections and circuitry of the brain at this level will help uncover the pathologies that underlie the common degenerative diseases of the brain, such as Alzheimer’s and Parkinson’s disease.
The work was funded by the British Neuropathological Society, Alzheimer’s Research UK and by an Innovation grant from Parkinson’s UK.
Images from the study were acquired using confocal microscope provided by the Imperial FILM Facility at the Hammersmith Campus.
-
‘Next generation histology methods for three-dimensional imaging of fresh and archival human brain tissues’ by Hai Ming Lai et al. is published in Nature Communications.
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Ryan O'Hare
Communications Division