Reading of biological molecules improved up to 100x by doubling the sensors
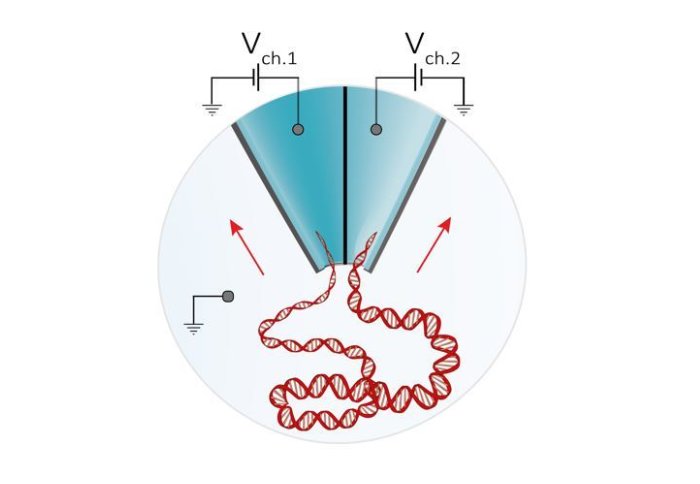
A coupled system of two miniature detectors called nanopores improves detection of biological molecules, including DNA and markers of early disease.
The ability to control the motion of single biological molecules is key to improving a wide range of biophysical and diagnostic applications, such as DNA sequencing and the detection of rare molecules linked to diagnosis and prognosis of disease.
The strength and reliability of the platform opens a plethora of possible applications. Professor Joshua Edel
In recent years, nanopores - tiny holes within insulating materials - have emerged as a promising tool capable of achieving these tasks. In nanopore sensing, individual molecules are passed through a very small nanometer-sized hole. This process results in each molecule producing a unique signature, without the need for lengthy sample preparation or chemical modification.
However, the smaller the molecule, the harder it is to detect. Very precise control is required to select single molecules and hold them in place long enough to be analysed.
In a collaborative effort, teams led by Dr Alex Ivanov and Professor Joshua Edel at Imperial College London and Professor Cees Dekker at the Delft University of Technology have developed a nanoscale sensor that holds molecules to facilitate a nearly 100x improvement in readout times.
The new technology, reported in the journal Nano Letters, works by actively controlling the transport of molecules in suspension using a double-nanopore architecture, where two nanopores are separated by a gap of approximately 20 nm wide (20 billionths of a meter). This effectively ‘traps’ molecules long enough for the nanopores to obtain accurate readouts.
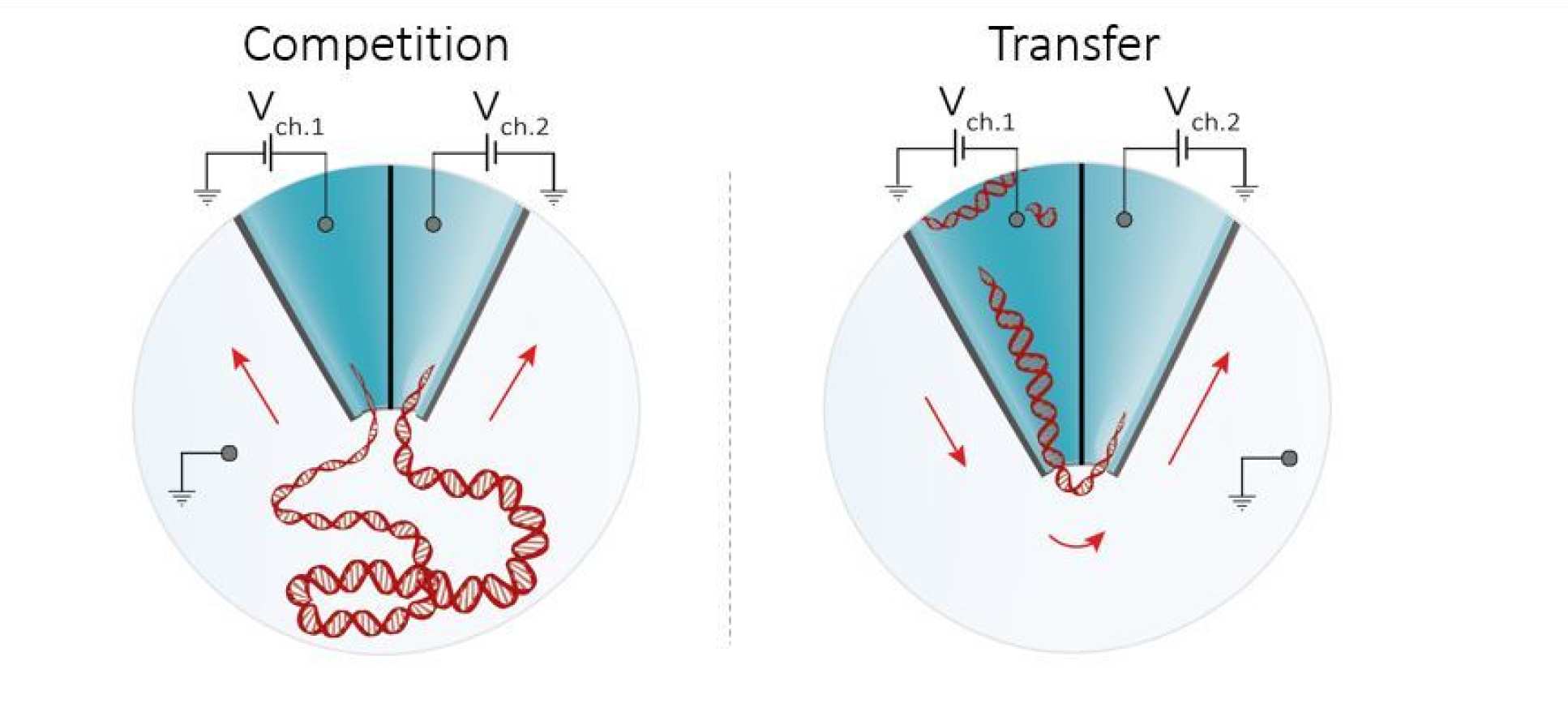
Co-leader of the study, Dr Ivanov, from the Department of Chemistry at Imperial, said: “Initially, both teams independently developed systems where two such nanoscale detectors are manufactured in close proximity. However, in the present study we combined the benefits of both methods to achieve significant improvement in the stalling of single molecules at the detector head.”
In addition, the method also allows precise control of molecular transport and the shuffling of single molecules from one detector to another with nearly 100% efficiency.
Professor Edel, also from the Department of Chemistry at Imperial, commented: “The strength and reliability of the platform opens a plethora of possible applications. For example, implementation of a feedback control mechanism would allow for better modulation and control of molecular transport.
“As an example, such a mechanism could be used to perform multiple readings of the same DNA molecule, yielding more accurate information about the molecules being probed. ”
Dr Ivanov added: “The whole project was only made possible by the enthusiasm of the young team members, including Paolo Cadinu, Giulia Campolo at Imperial and Sergii Pud at the Delft University of Technology, who all have diverse expertise and backgrounds. We were recently awarded an Imperial European Partners Fund grant to further develop our collaboration.”
-
‘Double barrel nanopores as a new tool for controlling single-molecule transport’ by Paolo Cadinu, Giulia Campolo, Sergii Pud, Wayne Yang, Joshua B. Edel, Cees Dekker and Aleksandar P. Ivanov is published in Nano Letters.
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Hayley Dunning
Communications Division