CRISPR‐TAPE: protein‐centric CRISPR guide design
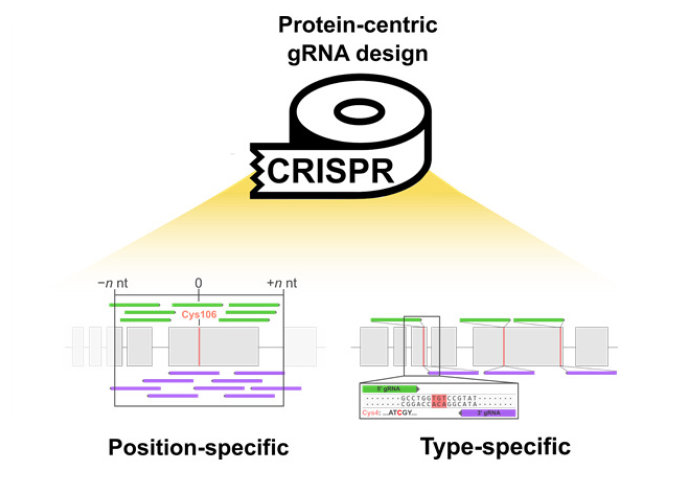
Protein‐centric CRISPR guide design has been developed for targeted proteome engineering
Congratulations to group member Henry Benns on his co-first authorship publication in Molecular Systems Biology.
The work was led by Dr Matt Child’s group in the Department of Life Sciences and reports the development of a new bioinformatics tool for protein-centric design of CRISPR guide RNAs (gRNAs).
CRISPR/Cas is a genome engineering technology that relies on the targeting of an RNA-guided endonuclease to introduce double-stranded breaks at specific genomic loci. One key application of CRISPR/Cas is site-directed mutagenesis, in which specific amino acids within protein-coding genes are modified through the integration of foreign DNA templates. To increase the efficiency of editing, gRNAs must target Cas9 in close proximity to the site of template integration. Despite this, search algorithms for the design of gRNAs have remained gene-centric, generating extensive lists of gRNAs that require time-consuming manual curation to identify those targeting specific protein regions of interest.
To address this, the team developed CRISPR-TAPE, a protein-centric gRNA design tool that allows users to identify guides that target specific residues or amino acid types within proteins. In CRISPR-TAPE, gRNA outputs are positionally filtered around an amino acid(s) of interest, enabling quick/automated identification of guides for downstream mutagenesis strategies. It is anticipated that the tool will make CRISPR-based protein engineering more accessible to the chemical biology community, empowering researchers seeking to modify specific amino acid chemistries.
CRISPR-TAPE is freely available from The Child Lab as a standalone Python script and as an executable application.
This work was funded by the BBSRC, The Royal Society, and Wellcome Trust.
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Dr Ravi Singh
Department of Chemistry