Bacterial warfare provides new antibiotic target
Pseudomonas aeruginosa bacteria
A detailed study of the weapons bacteria use to defeat their rivals has revealed a brand new target for antibiotics.
The finding could lead to a new class of antibiotics, as well as providing a method for finding other new targets by exploiting the tricks of bacterial warfare.
[Bacteria] have fought each other for billions of years and surely know the most effective targets to hit in order to kill competitors. Dr Laura Nolan
Antibiotic resistance, where pathogenic bacteria evolve resistance to drugs that usually kill them, is a rising problem globally, meaning new antibiotics need to be found. However, it is difficult for researchers to know which parts of bacterial cells to target with new drugs.
The new study, led by the team of Professor Alain Filloux at Imperial College London and published today in Nature Microbiology, has identified a new potential drug target that could be an ideal candidate for the development of novel antibiotics.
Asking bacteria
Lead author Dr Laura Nolan, previously from the MRC Centre for Molecular Bacteriology and Infection and now at the National Heart and Lung Institute at Imperial, said: “Despite constant efforts to develop new antibiotics, levels of antibiotic resistance continue to increase. While many avenues for the development of new drugs have been investigated, we’re still not sure what the best targets are for novel antibiotic therapies.
“We thought since bacteria can provide the answers, why not ask them directly? At the end of the day, they have fought each other for billions of years and surely know the most effective targets to hit in order to kill competitors.”
Different species, and even different strains of the same species of bacteria, can compete for resources by launching weapons that impair or kill their competitors. One of these weapons is the ‘Type VI secretion system’ (T6SS), a microscopic harpoon, which several bacteria use to fire toxic arrows at other bacteria.
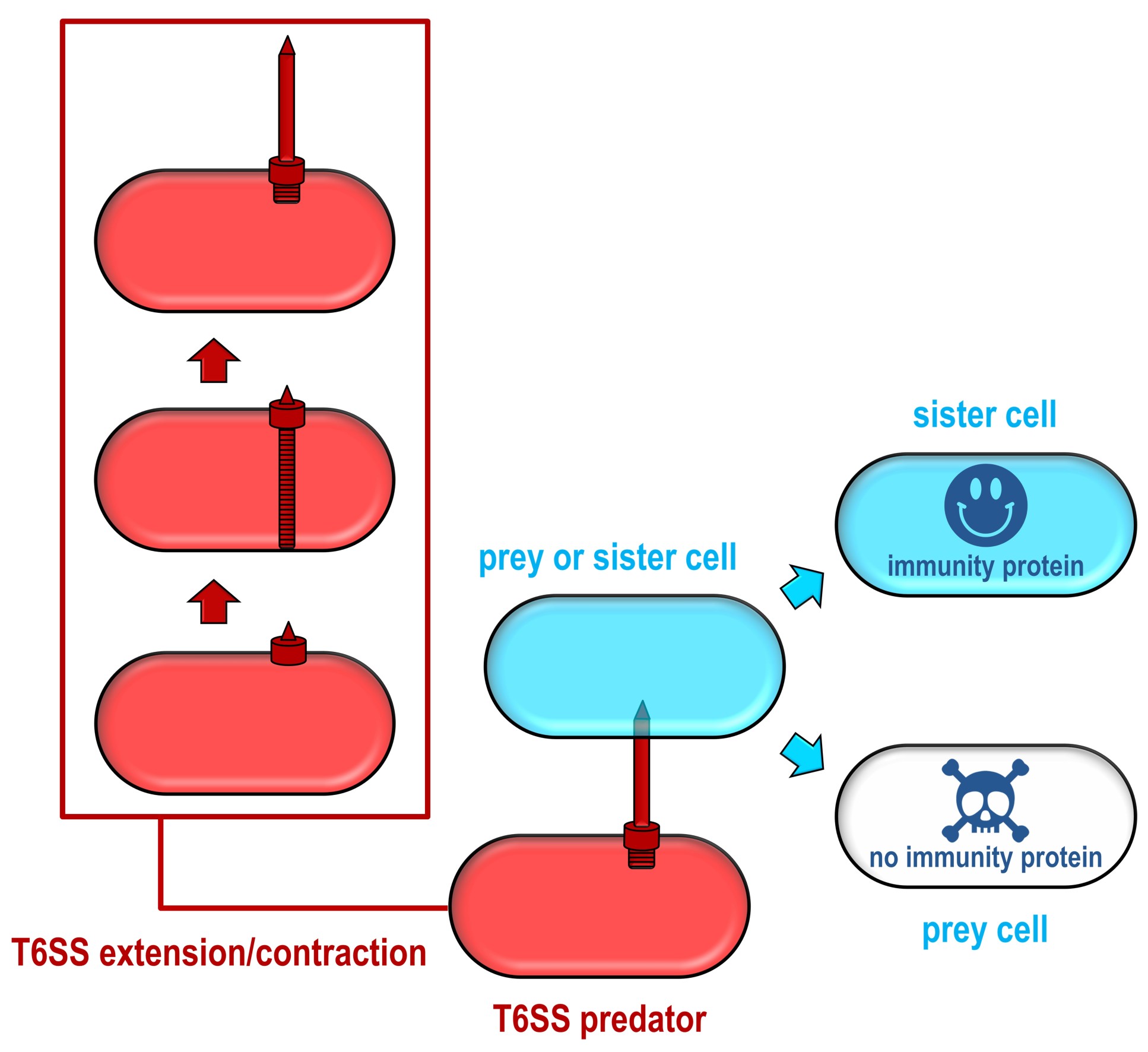
While several of these toxins are known, the researchers suspected other toxins, which attack different parts of their rival bacteria, were waiting to be discovered. However, it’s difficult to know what to look for when the mechanism of toxicity is unknown, so the team took a different approach.
Cataloguing transposons
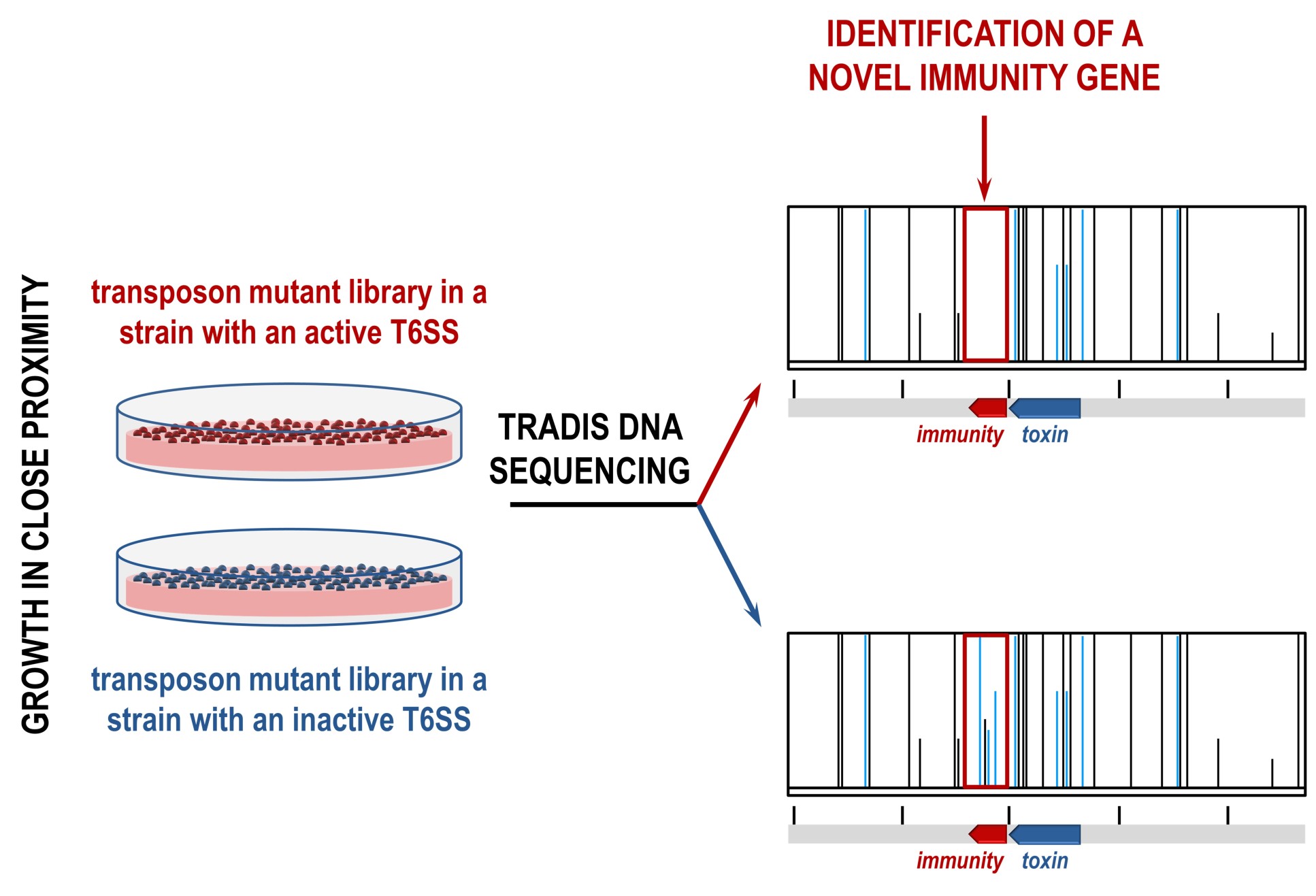
Every toxin gene sits next to a gene that encodes for an immunity against that toxin, which prevents cells from killing themselves or their neighbouring sister cells. The team realised that these immunity genes, and neighbouring toxins, could be identified using an approach called transposon directed insertion-site sequencing (TraDIS).
Transposons are DNA sequences that can change their position in the genome, often modifying genes. In this case, the team were looking for transposons that inactivated previously unknown immunity genes.
The team made two TraDIS libraries, each containing a pool of millions of bacterial cells that each contain unique transposon insertions across the whole genome. In one library, the T6SS was active and able to produce toxins, and in the other the T6SS was inactive and thus unable to produce toxins.
In this case, transposon insertions would only occur in the T6SS inactive library, allowing the team to find differences in the libraries. This led to the identification of previously known T6SS toxins and immunities, as well as a range of new ones.
They then studied one of the new toxin genes, known as Tse8, to find out how it was able to harm its competitors. They found that it impacted the functioning of a cell component called the transamidosome complex, which is crucial for protein synthesis in certain bacteria. Tse8 causes the transamidosome complex to ‘gum up’ by overly promoting the recruitment of one of its components, ultimately impairing the growth of the affected bacteria.
Professor Filloux said: “The identification of a toxin that impacts the transamidosome complex points to a new target for antibiotics, and knowing how the toxin works means we can now seek to find, or design, small molecules that could produce the same effect.”
Aiming at a new target
The transamidosome complex is also crucial for the survival of other pathogenic bacteria, including those that cause meningitis, urinary tract infections, and staph infections, meaning any antibiotic targeting it could have a range of uses. At the same time, the new antibiotic would not harm the so-called ‘good’ bacteria that live in our gut and support our health.
In addition, the method the team used could be replicated for finding new antibiotic targets that bacteria already exploit when fighting each other.
Co-corresponding author Dr Despoina Mavridou, previously at the MRC Centre for Molecular Bacteriology and Infection at Imperial and now at the University of Texas at Austin, said: “Our work makes a strong argument for the use of global genomics-based approaches for the identification of novel T6SS toxins, since it has the potential to uncover an extensive reservoir of unexplored yet naturally validated antibacterial targets.”
Next, the team will further characterise the mechanism of action of Tse8 to specify how the transamidosome complex could be chemically inhibited, as well as use the same unbiased approach to search for other potential new antibiotic targets within the T6SS.
-
‘Identification of Tse8 as a Type VI secretion system toxin from Pseudomonas aeruginosa that targets the bacterial transamidosome to inhibit protein synthesis in prey cells’ by Laura M. Nolan, Amy K. Cain, Thomas Clamens, R. Christopher D. Furniss, Eleni Manoli, Maria A. Sainz-Polo, Gordon Dougan, David Albesa-Jové, Julian Parkhill, Despoina A.I. Mavridou and Alain Filloux is published in Nature Microbiology.
Top image credit: Helga Mikkelsen
Article supporters
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Hayley Dunning
Communications Division