Nanopore sequencing and DNA barcoding method gives hope of personalised medicine
Credit: Jeroen Claus, Phospho Biomedical Animation
With the ability to map dozens of biomarkers at once, a new method could transform testing for conditions including heart disease and cancer.
Currently, many diseases are diagnosed from blood tests that look for one biomarker (such as a protein or other small molecule) or, at most, a couple of biomarkers of the same type.
The new method, developed by scientists at Imperial College London in a research collaboration with Oxford Nanopore Technologies (Oxford Nanopore), can analyse dozens of biomarkers of different types at the same time. This would potentially allow clinicians to gather more information about a patient's disease.
The ability to monitor different types of molecules at the same time, in the same sample, offers a distinct advantage over traditional analysis methods. Ben Reilly-O'Donnell
For example, current tests for heart failure look for a couple of common proteins to tell whether the condition is present. The new method was able to additionally detect 40 different types of miRNA molecules, which have the potential to be used as a new class of biomarkers. It can simultaneously examine proteins, small molecules like neurotransmitters, and miRNA from the same clinical sample, providing comprehensive data for a more precise diagnosis.
The results of using the new test in this way with the blood of healthy participants, for a proof-of-concept study, are published today in Nature Nanotechnology.

Co-first author Caroline Koch, from the Department of Chemistry at Imperial, said: "There are many different ways you can arrive at heart failure, but our test will hopefully provide a low-cost and rapid way to find this out and help guide treatment options. This kind of result is possible with less than a millilitre of blood. It's also a very adaptable method so that by changing the target biomarkers it could be used to detect the characteristics of diseases including cancer and neurodegenerative conditions."
Co-first author Ben Reilly-O'Donnell, from the National Heart and Lung Institute at Imperial, added: "The ability to monitor different types of molecules at the same time, in the same sample, offers a distinct advantage over traditional analysis methods."
DNA barcoding
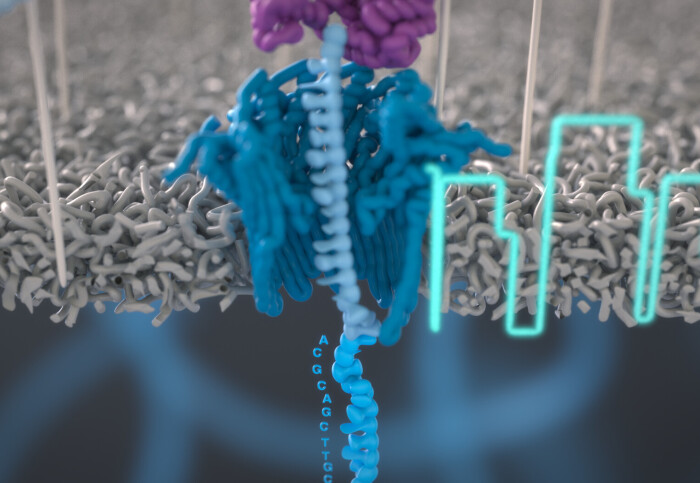
The test works by mixing the blood sample with DNA 'barcodes'. These are small tags made of short DNA sequences, each encoding a unique probe designed to attach to a different biomarker. Once the sample and barcodes have been mixed, the resulting solution is injected into a low-cost handheld device previously developed by Oxford Nanopore – the MinION.
The Oxford Nanopore device holds a flow cell, containing an array of nanopores – very small holes – that are able to read the electrical signature from each DNA barcode that passes through them. The complex electrical signal the device produces is interpreted by a machine-learning algorithm to identify the type and concentration of each biomarker present in the sample.

The DNA barcodes used for each test can be made to order, specifically for the biomarkers that need to be analysed to characterise the disease being tested for. Using DNA barcodes also removes the need for complex and time-consuming sample preparation, which can also introduce sample bias.
Lead researcher Professor Joshua Edel, from the Department of Chemistry at Imperial, said: "Working with Oxford Nanopore Technologies, we have been able to take their existing platform and innovate how it can be used, with the addition of DNA barcodes and machine learning to understand the results."
Co-lead researcher Dr Alex Ivanov, also from the Department of Chemistry at Imperial, said: "In principle, we are close to enabling a technology being suitable for clinics, where, in the long run, we hope it could provide a wealth of individualised information for patients with a range of conditions."
Future directions
After showing that this method can successfully measure 40 miRNA molecules in healthy patient blood, the team are now working with clinical samples from heart failure patients to validate the results. Regular testing like this could also help clinicians establish their individual patient baselines for common blood biomarkers.
The method may be useful for speeding up diagnosis in two ways: as well as measuring more biomarkers at once, it can also help find new biomarkers. While currently only a handful of biomarkers are validated for diagnosing heart disease, by measuring 40 miRNA types of interest simultaneously, the team could also see which of these are relevant and could be validated with more testing.
-
'Nanopore sequencing of DNA-barcoded probes for highly multiplexed detection of microRNA, proteins and small biomarkers,' by Caroline Koch et al., is published in Nature Nanotechnology.
Article supporters
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Hayley Dunning
Communications Division