Unusual RNA structures could be targets for new ALS treatments
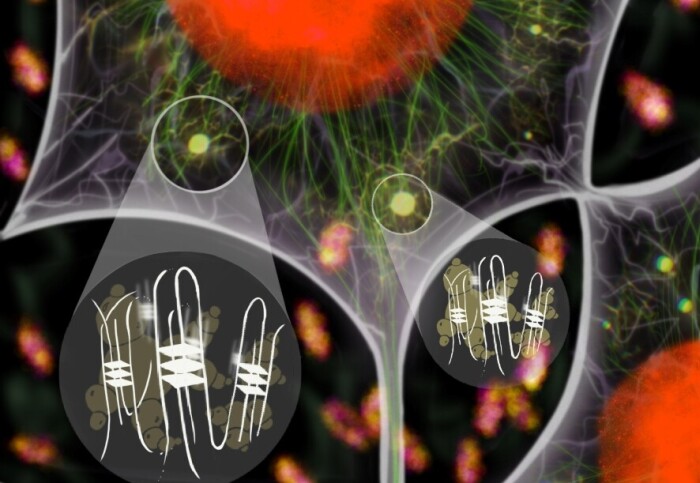
Studying strange forms of RNA associated with the formation of aggregates in the brains of ALS patients could lead to new avenues for treatments.
Amyotrophic lateral sclerosis (ALS) is a progressive motor neuron disease, which causes degeneration of nerve cells in the spinal cord and brain. Neurodegenerative diseases, including ALS, dementia, and Alzheimer’s, are the leading cause of death in the UK, and there are no known cures.
New therapies are desperately needed for these diseases, and drugs based on disrupting the accumulation of mG4s and other unusual RNA structures could play a key role in the future.” Dr Marco Di Antonio
As well as physical symptoms, one of the molecular hallmarks of these diseases is the formation of solid aggregates in the brains of sufferers, which have been thought to be mainly driven by certain proteins. Several drugs have been designed to break apart these protein aggregates, but none have so far achieved significant improvements in symptoms.
Now, a team led by Dr Marco Di Antonio at Imperial College London, in collaboration with Dr Lorenzo Di Michele (University of Cambridge) and Professor Rickie Patani (The Francis Crick Institute and UCL Queen Square Institute of Neurology), have looked at an alternative mechanism that could lead to aggregate formation in ALS, and found that unusual forms of RNA may play a key role in their build-up.
The findings could lead to new drugs that target these RNA structures, and points to the potential of searching for similar factors in related neurodegenerative diseases. The research is published in Nature Communications.
First author Federica Raguseo, who undertook the research for her PhD in the Department of Chemistry at Imperial, said: “For more than 20 years we have looked at protein aggregations as the cause of neurodegenerative diseases, but the poor performance of many candidate drugs suggests we might be missing something: that aggregates may be a symptom, rather than a cause.
“Looking at the causes of the aggregates may lead us to the causes of the symptoms themselves, which could help us find new ways to tackle these debilitating diseases.”
Four-stranded RNA
The Imperial researchers had recently discovered a human protein that can promote the formation of a special structure of DNA called multimolecular G-quadruplex (mG4), which is made of four separate strands of DNA instead of the usual two. These structures can connect distant parts of DNA strands, which is thought to help condense DNA in the nucleus of our cells.
Looking at the causes of the aggregates may lead us to the causes of the symptoms themselves, which could help us find new ways to tackle these debilitating diseases. Federica Raguseo
The team has now looked at condensation of RNA rather than DNA in the gene C9orf72, which is mutated and heavily expanded in some ALS cases. The gene contains a ‘repeat expansion’ – a repeating section in the DNA that is converted into RNA when the DNA is read.
By studying the behaviour of the C9orf72 RNA, the team discovered that ALS-related RNA expansions can generate mG4s, forming a network that leads to solid aggregates. These mG4 structures can aggregate in the absence of proteins, potentially provide a base for proteins to aggregate around.
To test this, the team showed how proteins were more likely to aggregate in the presence of mG4 networks, and that stopping the mG4-formation prevents RNA condensation and aggregation of proteins.
A new place to look
These tests were performed with molecules in the lab, so to check the same is likely to be true in people, the team then looked for mG4 aggregation in the spinal motor neurons of ALS patients. As expected, the tests showed accumulations of G4s within aggregates in the patient derived spinal motor neurons, a cell type that is principally affected in the disease.
The team next want to investigate the association between mG4s and protein aggregates in more complex real-world situations, to see if the cellular environment has any effect, and whether there are viable pathways to blocking mG4 aggregation in these situations.
Lead researcher Dr Marco Di Antonio, from the Department of Chemistry at Imperial, said: “What we have found is not the definitive answer to the puzzle, but we have shown that these unusual RNA structures likely contribute to formation of aggregates in ALS. This offers an alternative pathway for aggregate formation, which should be also looked in other neurodegenerative diseases.
“New therapies are desperately needed for these diseases, and drugs based on disrupting the accumulation of mG4s and other unusual RNA structures could play a key role in the future.”
-
‘The ALS/FTD-related C9orf72 hexanucleotide repeat expansion forms RNA condensates through multimolecular G-quadruplexes’ by Federica Raguseo et al., is published in Nature Communications.
Main image: Federica Raguseo
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Hayley Dunning
Communications Division