NAD+ precursor probes as tools for profiling intracellular ADP-ribosylation
Tate group researchers have performed the first in-depth metabolomics analysis with NAD+ precursor probes for profiling intracellular ADP-ribosylation
Past Tate group researchers Simeon Draganov and Michael Gruet, in collaboration with the group of Hector Keun, have published a study in RSC Chemical Biology reporting the use of a small molecule probe for profiling ADP-ribosylation protein targets in intact cells.
ADP-ribosylation, a type of post-translational modification found on proteins and nucleic acids, plays an important role in DNA damage repair, cell signalling and regulating protein function. Dysregulation of polyADP-ribosylating proteins (PARPs), responsible for ADP-ribosylation, has been linked to the development of several cancer types, such as breast and ovarian cancers. To elucidate the underlying mechanisms of dysregulation, novel tools are necessary to profile ADP-ribosylated proteins and PARP targets in a cellular context.
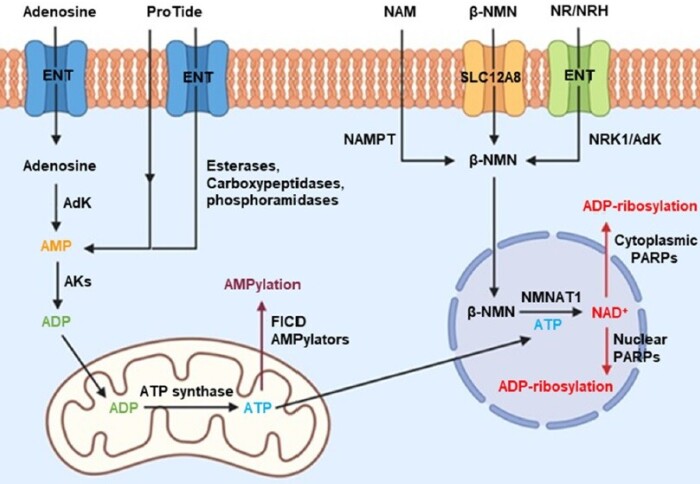
The study explores the use of click chemistry-based cell-permeable ProTide prodrugs, such as 6Yn-Pro, demonstrating that cell-permeable NAD+ analogue precursor probes are a viable option for profiling intracellular ADP-ribosylated targets. Through our in-depth metabolomics analysis of the NAD+ biosynthesis pathway we were able to detect conversion of 6Yn-Pro into the relevant metabolite analogues (6Yn-AMP, 6Yn-ATP, 6Yn-NAD+) intracellularly and quantify this conversion, both in absolute terms as well as relative incorporation into native metabolite pools. Our subsequent proteomics analysis confirmed that metabolically generated 6Yn-NAD+ was able to label known ADP-ribosylated proteins. We anticipate the described methodology and two novel ProTide probes, 2Yn-Pro and 8But-Yn-Pro, will serve as a basis for further optimisation of clickable NAD+ precursor probes for studying intracellular ADP-ribosylation events, including non-canonical ADP-ribosylation.
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Edward Bartlett
Department of Chemistry