Proximity-based labelling for proteomic mapping
Tate group PhD student Zuzanna Jablonska has published an editorial on proteomics mapping in Nature Reviews.
The new editorial describes how proximity-based labelling proteomics have emerged as a powerful tool to map the dynamic protein interaction networks relevant to physiology and pathology.
Conventional methods such as affinity purification and fractionation are often insufficient to capture transient protein-protein interactions. In the last decade, enzyme-catalysed proximity labelling has evolved to study these interactions both in vitro and in vivo. Utilising enzymes like APEX, BioID or TurboID, researchers have successfully identified and explored novel interactions, subcellular neighbourhoods and components of signalling pathways.
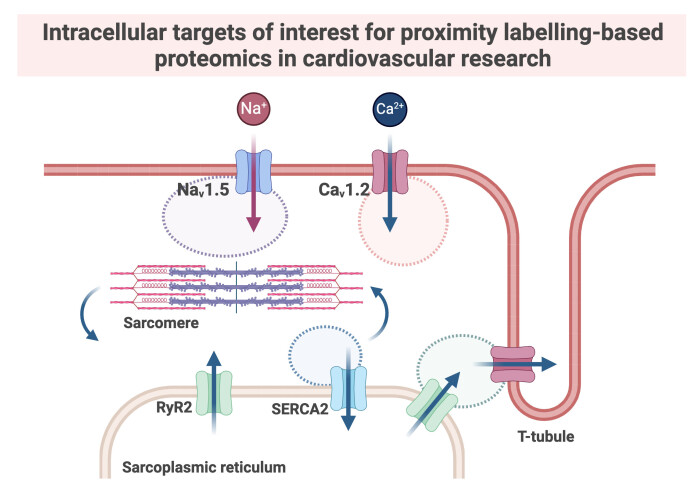
By genetically fusing enzymes, we can target specific proteins or compartments within cells and covalently tag proximal proteins. This allows for the enrichment of labelled proteins whilst preserving spatiotemporal dynamics, prior to mass spectrometry profiling. In cardiomyocytes, proximity-based proteomics was successfully used to map the interactomes of voltage gated channels CaV1.2 and NaV1.5. The data derived from proximity-based proteomics can improve our understanding of cardiac contractility and contribute to the identification of potential therapeutic targets for conditions like cardiac arrhythmias.
Congratulations on the publication Zuzanna!
Article supporters
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Edward Bartlett
Department of Chemistry