We describe High Content Analysis (HCA) to be the implementation of automated imaging for high throughput assays that include image acquisition with cellular or subcellular resolution. HCA is most commonly implemented using wide-field or confocal intensity imaging, often with multispectral detection, for which there is a range of available commercial instrumentation.
Our work applying advanced microscopy techniques, particularly FLIM/FRET, to biomedical studies highlighted the challenges to achieve reproducibility and statistical robustness when undertaking manual microscopy experiments, noting that the sample preparation could change between repeats of experiments and the acquisition and image analysis could be subjective and therefore open to operator-bias. Accordingly, we were motivated to implement our microscopy techniques in an automated multiwell plate imaging format. The consequent near-uniform sample preparation in each well and the opportunity to image 100’s of fields of view under similar conditions enables averaging over large sample numbers – typically with many replicates of the same experimental conditions and wells with control or calibration samples. This, combined with the high consistency of sample preparation across a multiwell plate, significantly reduces the impact of random noise in both measurement and biological systems (for example, expression of fluorescent proteins). The low random noise, consistent sample preparation and replicates of experimental conditions then enable any systematic errors in the measurement to be easily identified.
We are developing a modular open source HCA platform: openHCA, for MicroManager that will be extendable to multiple imaging modalities. This will eventually incorporate our existing HCA modalities:
High content analysis
Our first HCA experiments were motivated by the need to improve reproducibility of FLIM/FRET experiments and we developed an automated multiwell plate FLIM microscope that provided optical sectioning via a spinning disc (Yokogawa CSU-10 or Csu-X0 and FLIM via time-gated imaging using a GOI [1]. This was combined with FLIMfit [2] which can rapidly analyse FLIM data from 100’s of fields of view, including using global analysis. The operation of this FLIM HCA system has been describe in detail in a JovE article [3].
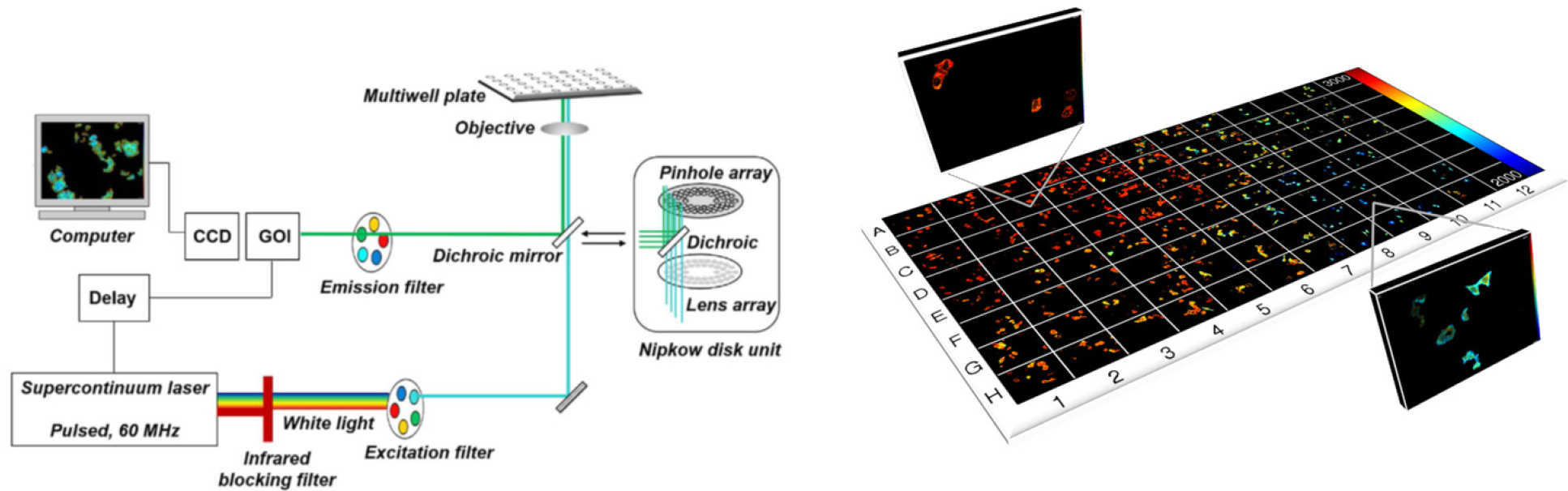
FLIM HCA has been applied to a range of assays for medical research, e.g. of HIV Gag protein aggregation [4], where dose response curves with Z’>0.5 were realised, and to RASSF signaling [5] where the minimum lifetime change required for a FLIM/FRET assay with plasmid-expressed fluorescent proteins was less than 20 ps and where we used global fitting to a complex decay model and measurements of fluorophore brightness to obtain, in situ, the dissociation constant, KD, for this quantitative cell-based assay. Most recently we have applied FLIM HCA to a FRET assay of interactions of endogenous yeast cell kinetochore proteins and RDH54 labelled with fluorescent proteins [6].
Super-resolved microscopy techniques are notoriously sensitive to the specific details of the sample preparation and so making comparisons between different experiments can be challenging. This suggests that HCA could add significant value to SRM-based studies, and it is interesting to explore readouts of super-resolved cellular structure as readouts of different phenotypes in drug discovery.
Our easySTORM approach [7] to SRM inherently provides large fields of view, allowing many cells to be studied in a single acquisition and the relatively simple and robust configuration can be implemented with automated multiwell plate imaging. We are developing this to study changes in cellular ultrastructure in chronic obstructive pulmonary disorder (COPD) in order to understand why some macrophages fail to phagocytose bacteria, fungi or other insults. Although STORM based assays are inherently slow due to the requirement to acquire 1000’s of images per field of view, the automated multiwell plate approach is probably the only practical way to acquire sufficient SRM data for systematic studies or assays. However, this easySTORM HCA results in a very significant data processing challenge. To address this, we utilise the parallelised SMLM data processing on a high-performance computing (HPC) cluster [8] using ThunderSTORM or other SMLM processing plug-ins.
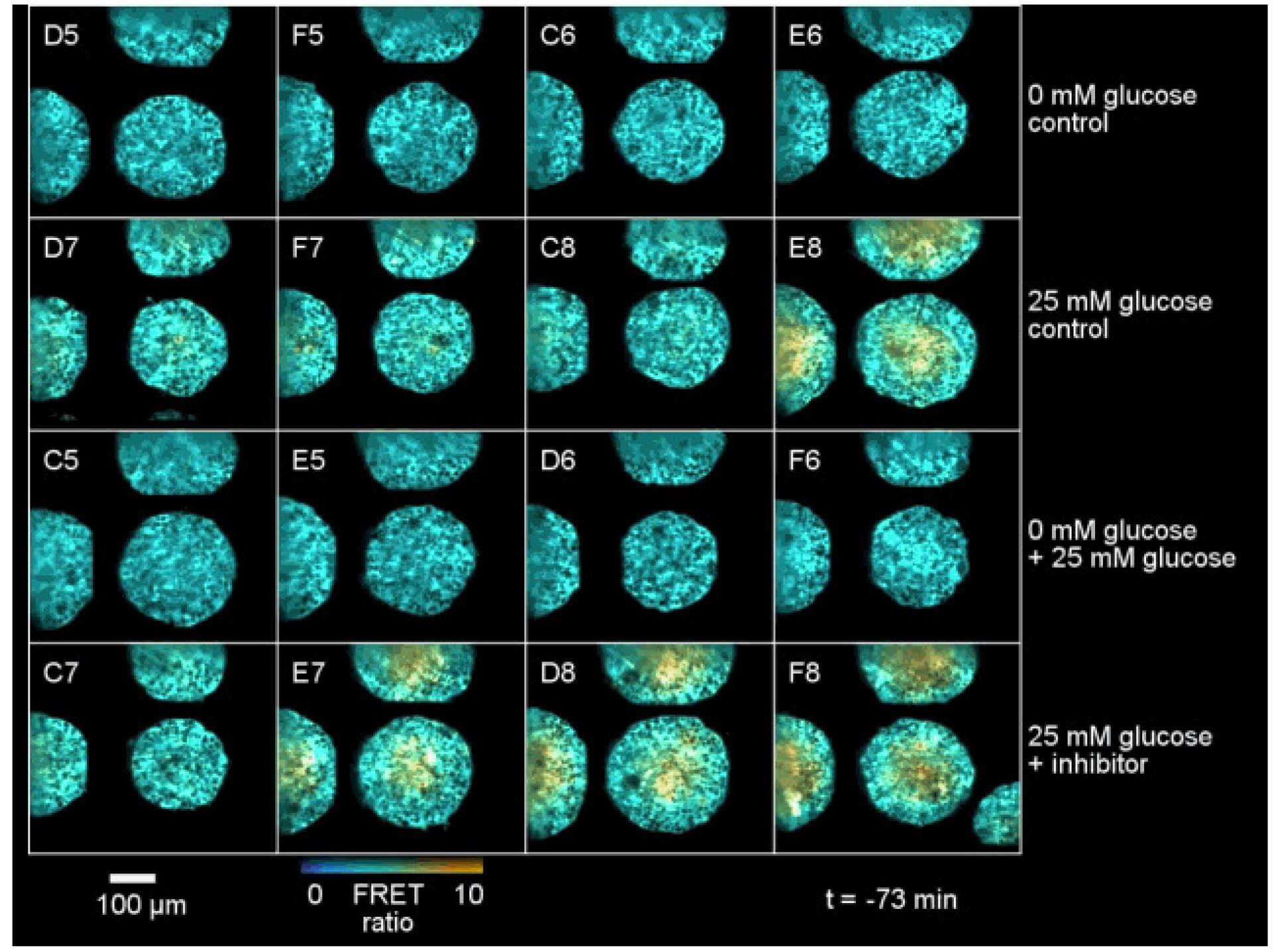
There is a rapidly increasing trend for cell biology to be studied in 3D spatial dimensions – with a shift from “2D cell cultures” (i.e. monolayers or cells on a coverslip) to 3D cell cultures, such as organoids or co-cultures in tissue matrix. Unfortunately, for cell biology studies and for drug discovery, this makes the imaging much more challenging – particularly for HCA where it is important to achieve a reasonable throughput to enable assays of 100’s of samples. The commercial state-of-the-art is multiwell plate spinning disc confocal microscopy, which provides rapid optical sectioning and slower acquisition of z-stacks for the acquisition of 3D data sets. We have addressed this using OPM to provide rapidly scanned volumetric imaging (up to ~30 vol/s) such that 3D time lapse live cell imaging in multiple wells is possible [9]. The figure below shows OPM HCA data for a spectrally-resolved FRET reading out a FRET biosensor for glucose expressed throughout tumour spheroids.
High Content analysis references
- High speed unsupervised fluorescence lifetime imaging confocal multiwell plate reader for high content analysis
C. B. Talbot, J. McGinty, D. M. Grant, E. J. McGhee, D. M. Owen, W. Zhang, T. D. Bunney, I. Munro, B. Isherwood, R. Eagle, A. Hargreaves, M. Katan, C. Dunsby, M. A. A. Neil and P.M. W. French
J. Biophoton. 1 (2008) 514–521 - Rapid global fitting of large fluorescence lifetime imaging microscopy datasets,
S.C. Warren, A. Margineanu, D. Alibhai, D.J. Kelly, C. Talbot, Y. Alexandrov, I. Munro, M. Katan, C. Dunsby and P.M.W. French, PLoS ONE 8 (2013) e70687 - Open Source High Content Analysis Utilizing Automated Fluorescence Lifetime Imaging Microscopy
F. Görlitz*, D. J. Kelly*, S. C. Warren, D. Alibhai, L. West, S. Kumar, Y. Alexandrov, I. Munro, J. McGinty, C. Talbot, R. A. Serwa, E. Thinon, V. da Paola, E. J. Murray, F. Stuhmeier, M. A. A. Neil, E. W. Tate, C. Dunsby and P. M. W. French
J. Vis. Exp. 119, (2017) e55119 - Automated fluorescence lifetime imaging plate reader and its application to Förster resonant energy transfer readout of Gag protein aggregation
D. Alibhai, D. J. Kelly, S. Warren, S. Kumar, A. Margineanu, R. A. Serwa, E. Thinon, Y. Alexandrov, E. J. Murray, F. Stuhmeier, E. W. Tate, M. A.A. Neil, C. Dunsby and P. M.W. French,
J. Biophotonics 6 (2012) 398-408 - Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM)
Anca Margineanu, Jia Jia Chan**, Douglas J. Kelly**, Sean C. Warren**, Delphine Flatters, Sunil Kumar, Matilda Katan, Christopher W. Dunsby, Paul M.W. French
Sci Rep. 6 (2016) 28186 - Automated FLIM HCA of protein-protein interactions between endogenously-labelled kinetochore proteins in live budding yeast cells
Wenjun Guo, Sunil Kumar, Frederik Görlitz, Edwin Garcia, Yuriy Alexandrov, Ian Munro, Douglas J. Kelly, Sean Warren, Peter Thorpe, Christopher Dunsby*, Paul French*
SLAS Technology, 24 (2019) 308-320 - easySTORM: a robust, lower-cost approach to localisation and TIRF microscopy
K. Kwakwa, A. Savell, T. Davies, I. Munro1 S. Parrinello, M.A. Purbhoo, C. Dunsby, M.A.A. Neil and P.M.W. French
J. Biophotonics 9 (2016) 948–957 - Accelerating single molecule localisation microscopy through parallel processing on a high-performance computing cluster HPC STORM, I. Munro*, E. García1*, M. Yan*, S. Guldbrand, S. Kumar, K. Kwakwa, C. Dunsby, M.A.A. Neil# and P.M.W. French#, J Micros. 273 (2019) 148-160
- Time-lapse 3-D measurements of a glucose biosensor in multicellular spheroids by light sheet fluorescence microscopy in commercial 96-well plates.
Vincent Maioli, George Chennell, Hugh Sparks, Tobia Lana, Sunil Kumar, David Carling, Alessandro Sardini & Chris Dunsby
Sci Rep 6, (2016) 37777